Development and application of a genetic baseline to inform bull trout passage and conservation
Hargrove, J.S., Delomas, T.A., Campbell, M.R. et al. Development and application of a genetic baseline to inform bull trout (Salvelinus confluentus) passage and conservation in the Snake River. Conserv Genet (2024).
Bull trout conservation in Hells Canyon | Idaho Fish and Game
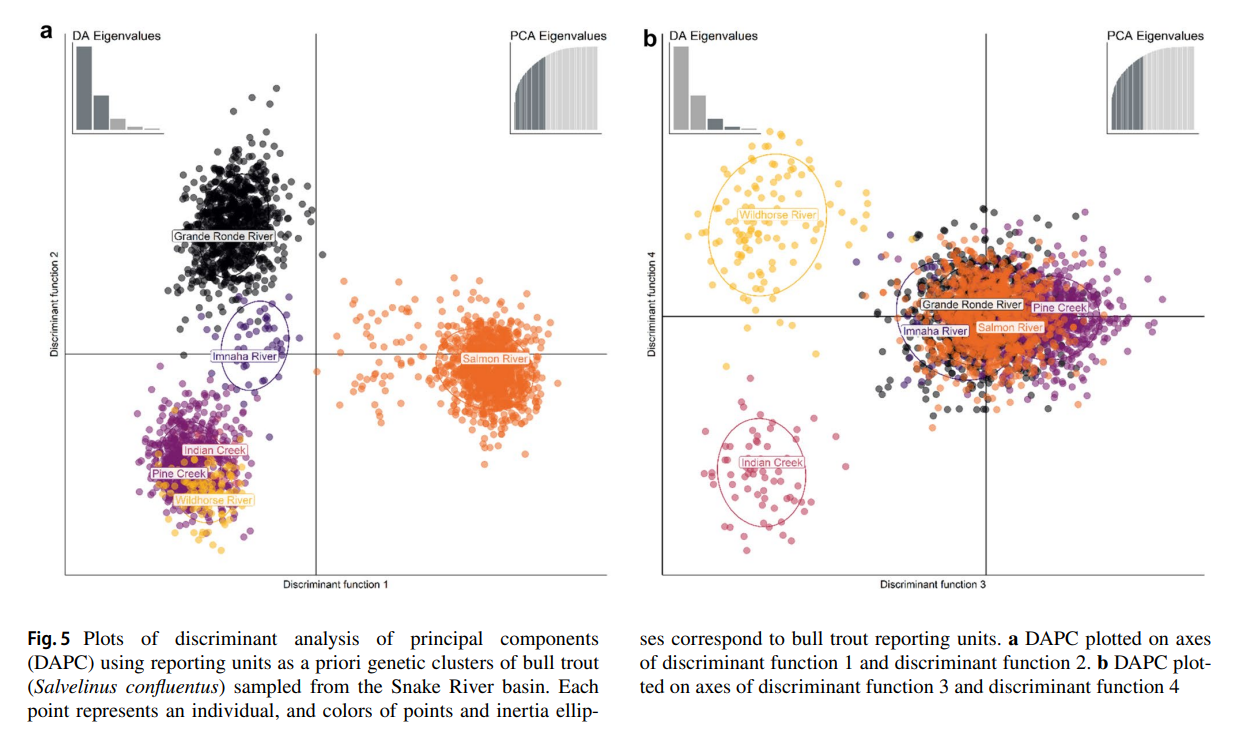
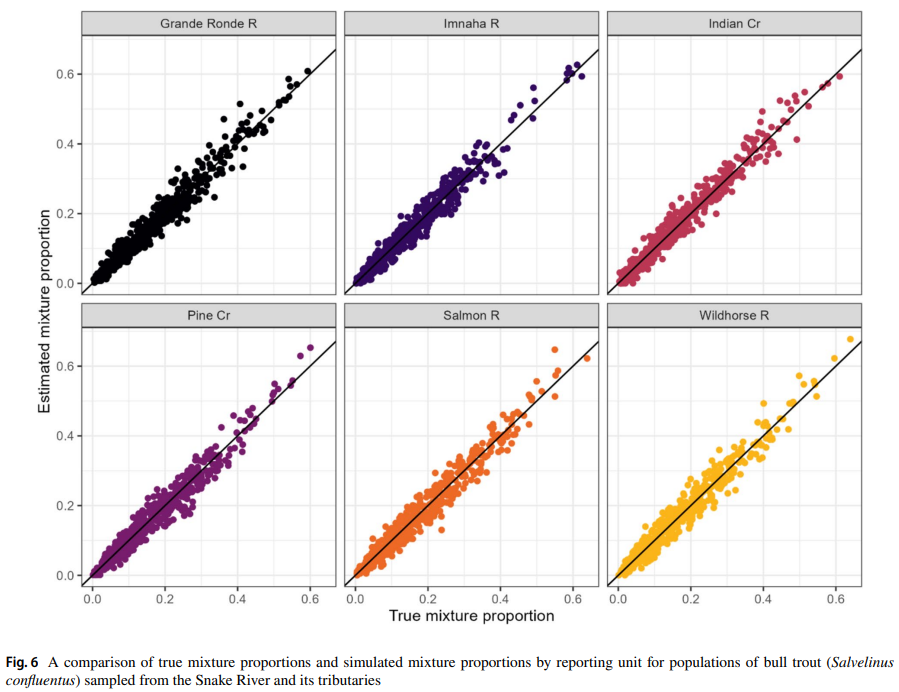
Barriers to upstream migration, such as dams, limit fish dispersal in lotic systems, including the federally protected bull trout (Salvelinus confluentus). The Hells Canyon Dam on the Snake River currently prevents upstream passage of migratory bull trout into tributary systems in Idaho and Oregon, USA. Viability modeling previously identified the influx of a small number of migrant bull trout as the most important variable for increasing population persistence of resident bull trout populations occupying headwater tributaries above Hells Canyon Dam. Migratory bull trout that overwinter in the Snake River below Hells Canyon Dam could potentially serve as a source of migrants via upstream passage; however, the origin of these fish and their genetic relationship to populations above Hells Canyon Dam is unknown. To address these knowledge gaps, we genotyped bull trout from Idaho and Oregon and developed a genetic baseline to identify the population origination of migratory bull trout below Hells Canyon Dam. An assessment of genetic baseline performance found that stock assignments were highly accurate and minimally biased. Genetic stock identification assigned 97% of the migratory bull trout captured below Hells Canyon Dam to the Imnaha River basin, a large bull trout population located below Hells Canyon Dam. No bull trout were assigned to populations above Hells Canyon Dam. Moving forward, the bull trout genetic data described here can serve as a baseline for future monitoring and conservation efforts in the transboundary waters of Oregon and Idaho and help inform select bull trout passage and translocation efforts.

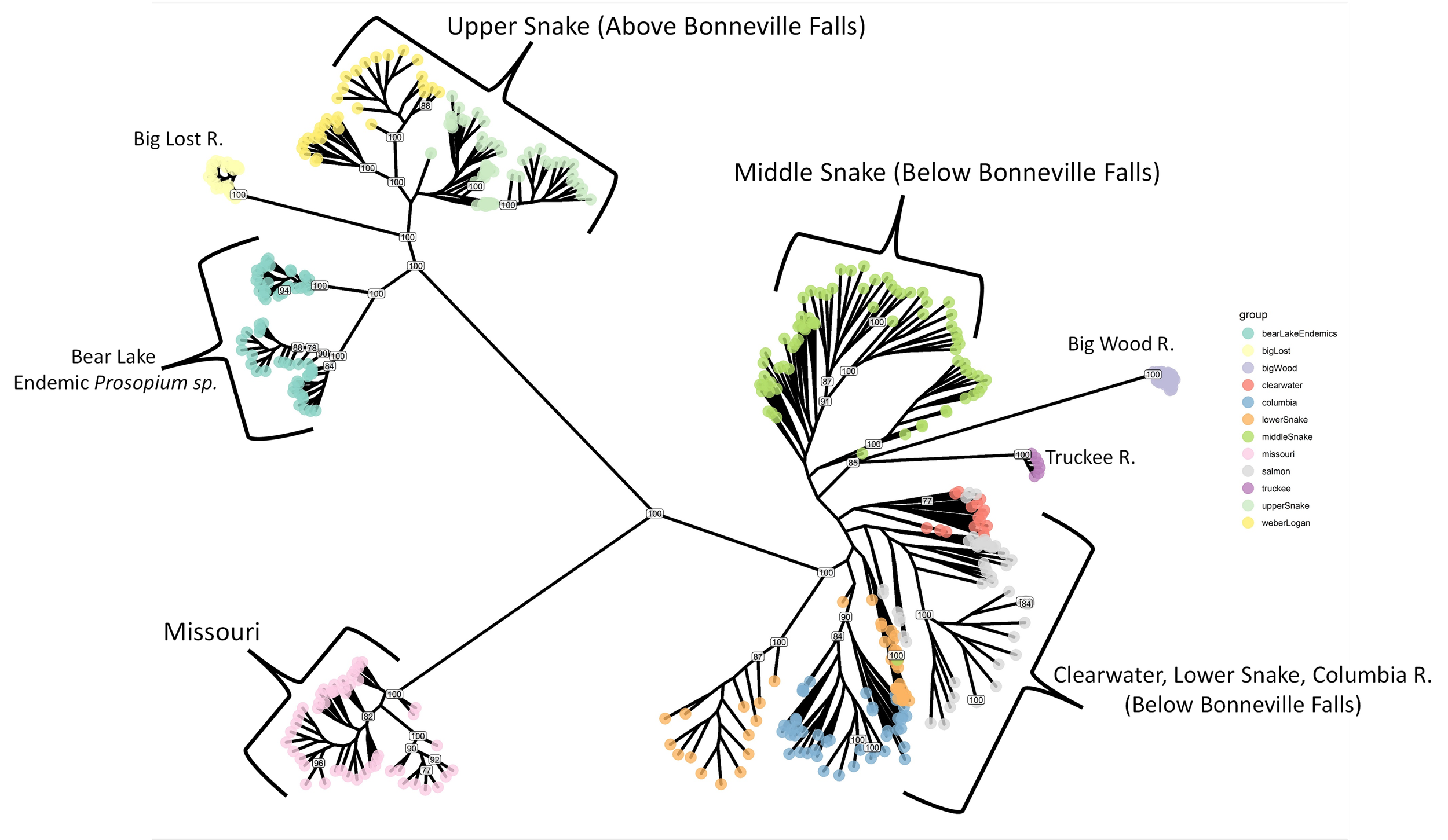
Mountain Whitefish Phylogeography
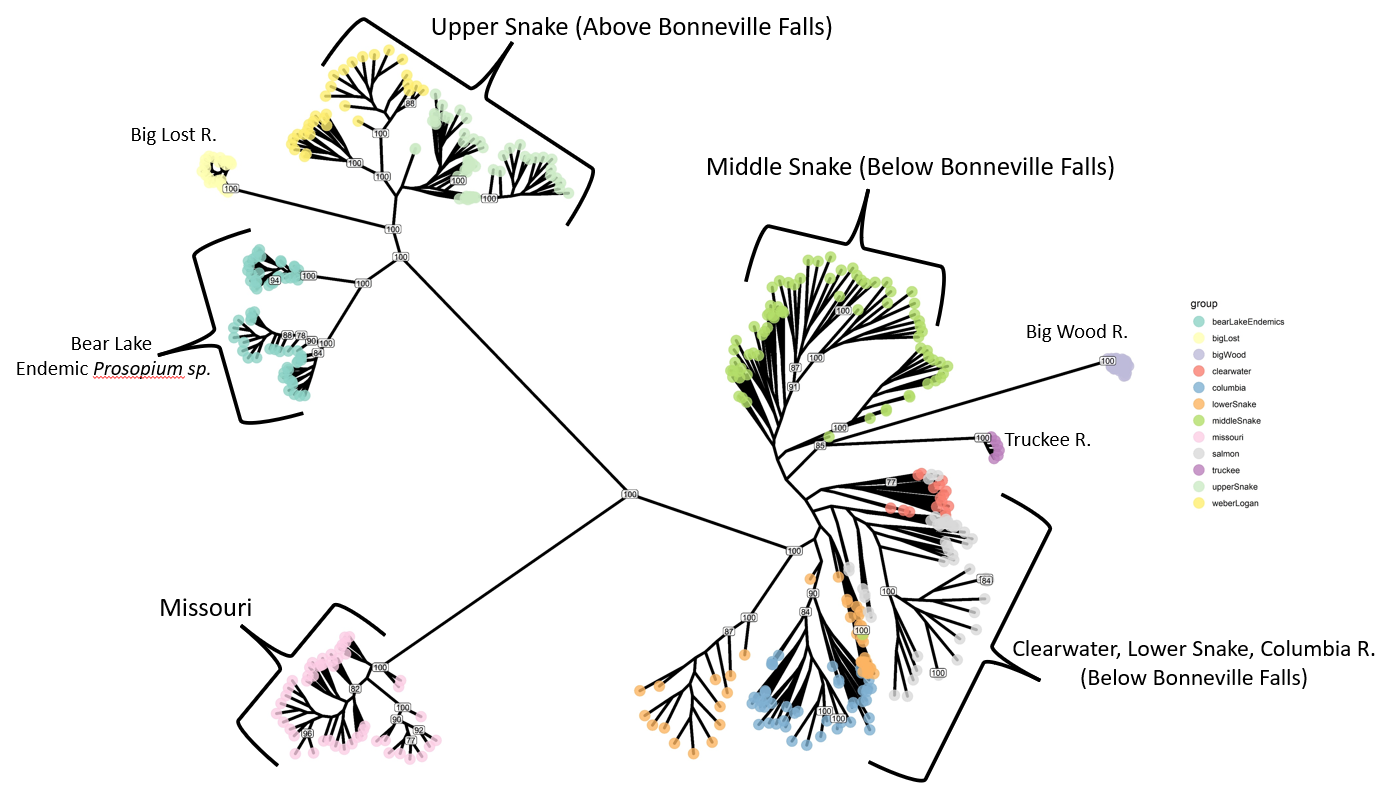
Mountain whitefish Prosopium Williamsoni are a widely distributed member of the Salmonidae family that are found throughout Western North America from the Lahontan Basin in California and Nevada, north to the Yukon-British Columbia Border in Canada. Past microsatellite and allozyme investigations have focused on understanding range-wide patterns of genetic structure and phylogeography of the species, with one goal being to identify and conserve historically isolated, genetically and evolutionarily independent, groups of populations. Despite this past research, no studies have been published that adequately describe the genetic structure of mountain whitefish throughout Idaho. In this study, we use restriction-site associated DNA sequencing and focus efforts on two river basins in central Idaho: Wood River and Lost River. These two basins are geographically adjacent and share nearby watersheds (within several kilometers from one another). However, by including Mountain Whitefish samples (and other Prosopium species) from throughout Idaho and adjacent states, we show definitively that these drainages were colonized by two separate ancient invasions. We discuss the conservation and management implications of these findings.
Project Leads: Katharine Coykendall and Audrey Harris


Snake River Sockeye Monitoring & Evaluation
Principal Biologist: Dave Venditti
Project Leaders: Eric Johnson, eric.johnson@idfg.idaho.gov and Odbayar Tumendemberel, odbayar.tumendemberel@idfg.idaho.gov
The Snake River Sockeye Salmon Captive Broodstock program was initiated in 1991 as a safety measure to avoid extinction of the stock. The research, monitoring and evaluation component of the program has been evaluating the contribution that hatchery produced Sockeye Salmon make towards the recovery of the ESU at several life stages (e.g. eyed eggs, presmolts, full term smolts, and adult releases). Field data are used to estimate juvenile and adult survival, the success of the different release strategies, and generate viable salmonid population parameters such as abundance, productivity, spatial structure and diversity. The research, monitoring and evaluation program also utilizes genetic tools and techniques to assist with broodstock collection and spawning and to measure effective population size and genetic diversity through time.
Putting the Red Back in Redfish Lake:
https://fisheries.org/docs/wp/AFS-Fisheries-November-2014.pdf#page=11
Genetic Diversity in the Snake River sockeye salmon captive broodstock program:





Genetic Stock Identification of wild steelhead and Chinook salmon in the Snake River basin
BPA funded project (2010-026-00).
Project Leaders: John Hargrove, john.hargrove@idfg.idaho.gov
NOAA recommends that to demonstrate ESU/DPS viability, annual documentation of a variety of biological parameters (including adult and juvenile abundance, age distribution, sex ratio, length, and run timing) is needed at the population, major population grouping, and ESU/DPS level for each listed species. This collaborative project with the Columbia River Inter-Tribal Fish Commission, utilizes Genetic Stock Identification (GSI) of Snake River steelhead and Chinook salmon adults and juveniles at Lower Granite Dam to provide estimates of these VSP parameters annually at the MPG and ESU/DPS level. GSI uses multi-locus genotype data from reference populations (presumably representing all contributing stocks) as a baseline and a complimentary set of genotype data from mixtures of fish of unknown origin to estimate stock proportions within the mixture. This project also provides several measures of genetic diversity and life history characteristics for adult steelhead and Chinook salmon detected at PIT tag arrays in the Snake River basin.
This project works directly with the Idaho Natural Production Monitoring and Evaluation Program (1990-055-00) and the Idaho Steelhead Monitoring and Evaluation Program (1991-073-00) to estimate the abundance and composition of wild adult steelhead and spring-summer Chinook salmon returning to the Snake River basin by body size (steelhead only), age, gender, and stock.
Important additional collaborators on this work includes:
-Nez Perce Tribe
-Idaho Office of Species Conservation (IOSC)
-Northwest Power and Conservation Council (NPCC)
-Pacific States Marine Fisheries Commission (PSMFC)
-Quantitative Consultants, Inc. (QCI)
-U. S. Fish and Wildlife Service
-Lower Snake River Compensation Program (LSRCP



Integrated Broodstock Status and Trend Monitoring
Principal Biologist: Dave Venditti
Project Leaders: Darcy McCarrick, darcy.mccarrick@idfg.idaho.gov and Odbayar Tumendemberel, odbayar.tumendemberel@idfg.idaho.gov
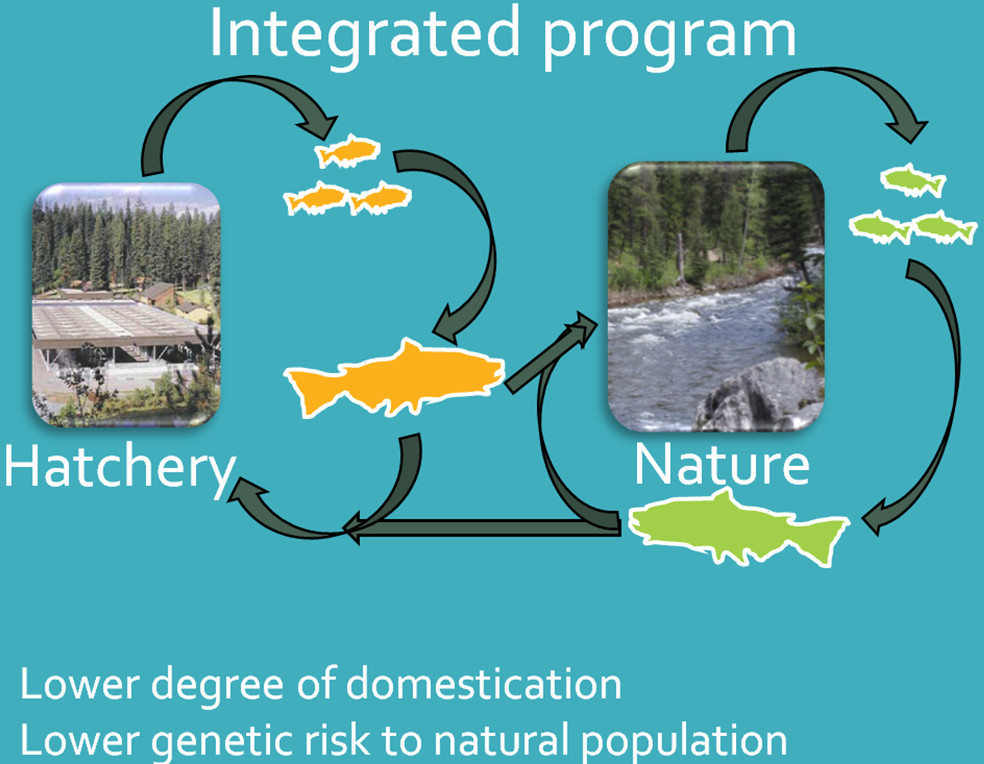
Supplementation programs can be implemented in a variety of ways, but all use hatchery production to augment naturally spawning fish populations. Supplementation programs have been implemented throughout the Columbia River basin for several decades (ISAB 2003). Recently, integrated broodstock (IB) programs have been developed in an effort to minimize genetic divergence between hatchery donor stocks and recipient wild stocks. A properly integrated program is one where the natural environment drives the adaptation and fitness of a composite population of fish spawning in a hatchery and in the wild (HSRG 2009). This is hypothesized to minimize domestication selection in the hatchery was well as ecological risks to the natural population (HSRG 2009). Integrated broodstock programs may also be used to increase the abundance of natural populations (Sharma et al. 2006; Berejikian et al. 2008), provide harvest opportunities (Fast et al. 2015), minimize risks to wild populations from hatchery straying (Mobrand et al. 2005), provide genetic repositories for natural fish in the hatchery environment (Kline and Flagg 2014), and may expand the distribution of fish spawning in under-utilized habitats (Dittman et al. 2010; Venditti et al. 2015).
This project will: 1) monitor the influence of natural spawners incorporated into integrated broodstocks on hatchery survival, productivity and replacement rates (HRR), and 2) monitor the influence of IB hatchery spawners on naturally spawning populations of Chinook Salmon (Oncorhynchus tshawytscha) and natural replacement rates (NRR).
This information will provide managers with a more complete picture of the benefits and risks of implementing IB programs in Idaho, which can guide the implementation of future supplementation strategies for Idaho and other populations in the Columbia River Basin.



Identification of SNPs for PBT of Burbot Lota Lota Using RAD Sequencing
Identification of SNP Markers for Parentage Based Tagging of Burbot Lota Lota Using Restriction-Site-Associated DNA Sequencing
Matthew R. Campbell1, Ryan Hardy1, Amanda Matala2 and Shawn R. Narum2, (1) Idaho Department of Fish and Game, (2) Columbia River Inter-Tribal Fish Commission
Project Summary:
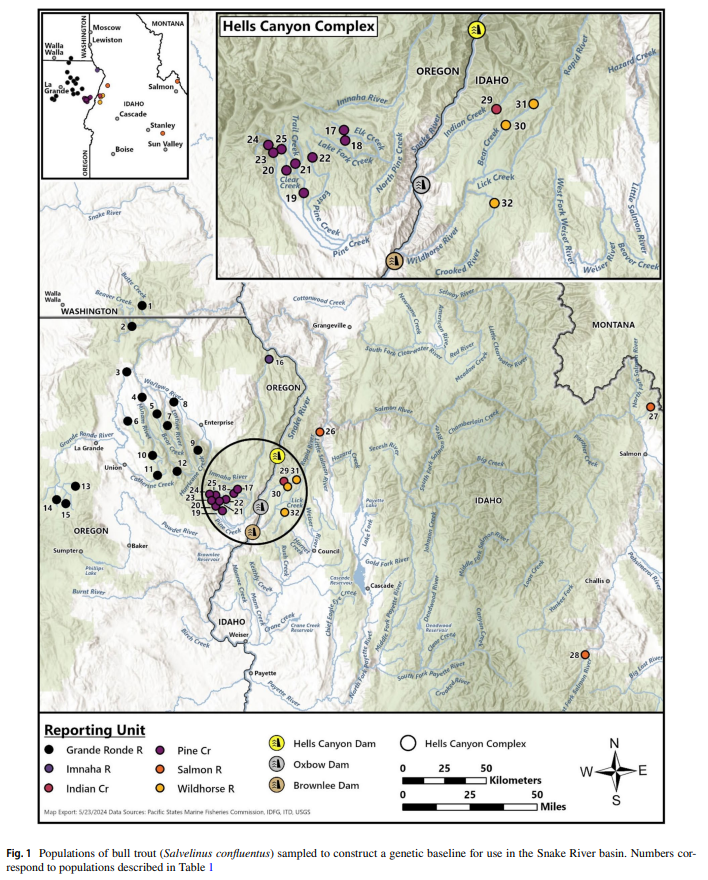
The trans-boundary (Idaho, USA and British Columbia, Canada) population of burbot lota lota native to the Kootenai River once provided a popular sport and commercial fishery, and is culturally significant to the Kootenai Tribe of Idaho. However, the population has experienced significant declines over the last 30 years, due primarily to habitat loss and alteration caused by water storage and diversion. By the late 1990’s, the population was considered functionally extinct, with estimates of fewer than 50 burbot in the wild and little to no recruitment, prompting an on-going international recovery effort. As part of these recovery efforts, managers have been actively developing a hatchery supplementation program to re-build the population and support future tribal sustenance harvest and recreational fisheries.
Each February, spawning adult burbot from Moyie Lake, B.C. are captured via angling, genetically sampled, and spawned on site (Figure 1). Fertilized eggs are transported to hatchery facilities for incubation and rearing. Juveniles are released for supplementation purposes in the Kootenai River, ID (Figure 2). While supplementation breeding programs have the potential to rapidly rebuild depleted natural populations, careful genetic management is critical. In order to monitor genetic diversity and potential inbreeding in the broodstock and to provide Parentage Based Tagging (PBT) of supplementation offspring, a powerful set of new genetic markers were needed. For this project we chose to utilize Restriction site associated DNA sequencing (RADSeq). This is a relatively new and cost-efficient technology allows the rapid discovery of thousands of SNP markers in species that have not been extensively studied previously or for which little existing DNA sequence data exists.
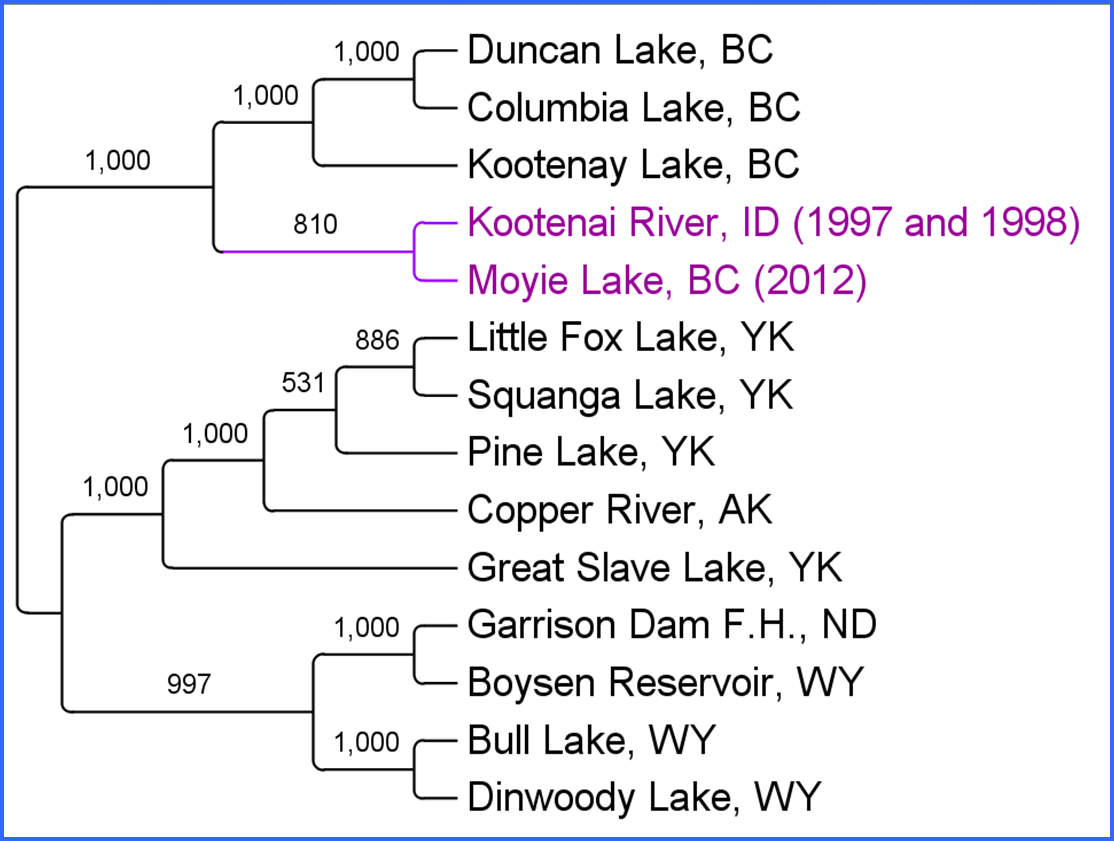
Using RADSeq technology, we discovered 6,517 SNPs and successfully genotyped 1,690 SNPs variable in Moyie Lake samples (Figure 3). The 1,690 SNPs were ranked by MAF (diversity) and HWE and 100 SNPs were submitted for assay-by-design (taqman genotyping assays; Figure 4). A total of 93 SNP assays identified for further genotyping. These exhibited ~44% HE in samples from Moyie Lake. We demonstrated the accuracy of our SNP set for parentage by running a known parent/offspring dataset (N = 344/313, respectively). All 313 offspring accurately assigned to the correct parent pair.
The SNP marker set provides a powerful new tool for managing broodstock and for monitoring and genetically tagging Burbot to track growth, post-release survival and movement of released individuals. In spawn year 2015, no crosses among full-siblings were detected and the effective population size of the spawning population was estimated as 397 (95% C.I. = 260 – 729). A total of 593 burbot adults were sampled from the Kootenai River in 2015. These samples were assigned back to a parental baseline consisting of all Moyie Lake parental broodstock spawned in 2011-2014 (N = 344). Of the 593 adults, we successfully assigned 497 (83.8%) back to the supplementation program (BY2011 = 377, BY2012 = 106, BY2013 = 14).

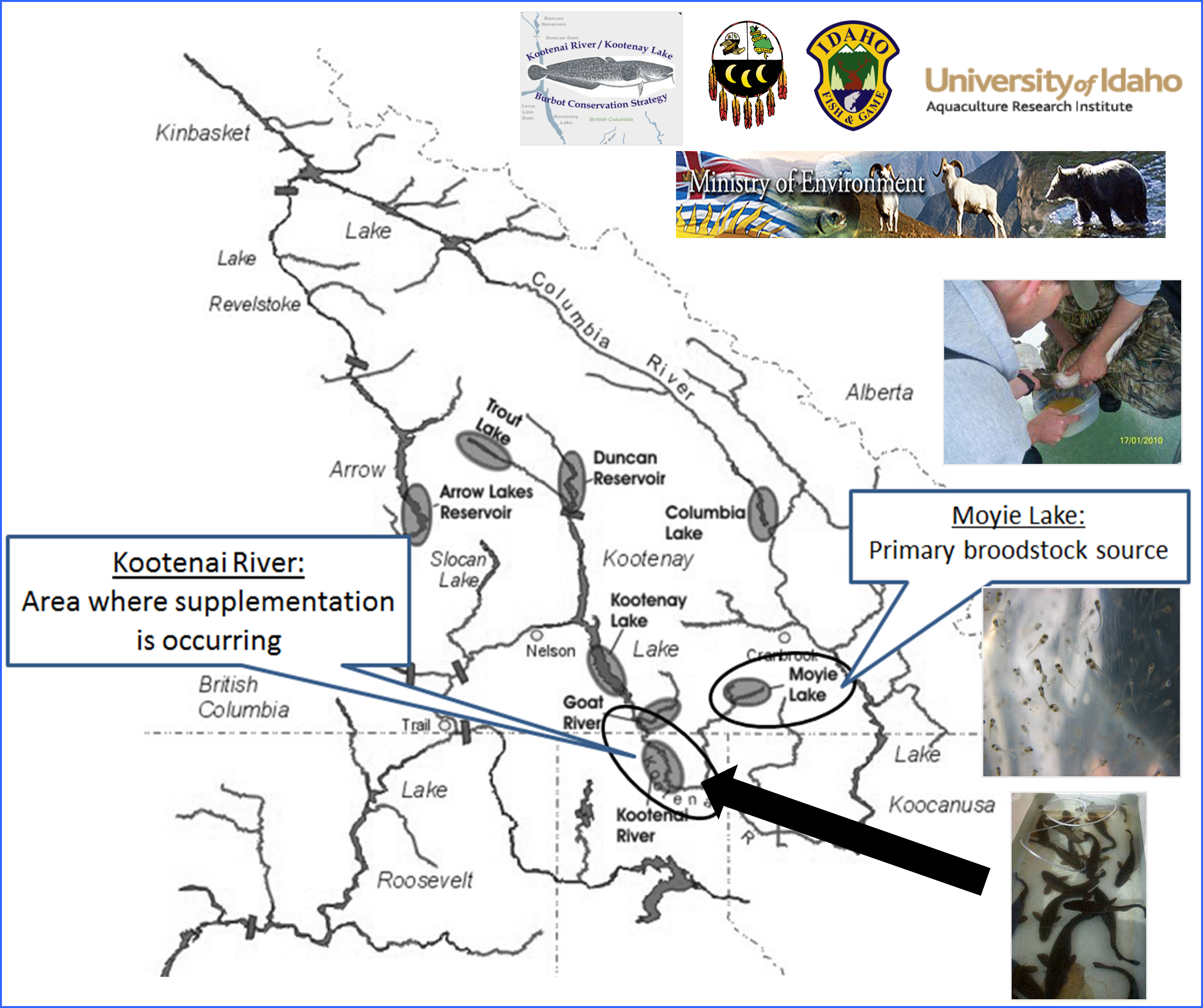
Each February, spawning adult burbot from Moyie Lake, B.C. are captured via angling, genetically sampled, and spawned on site. Fertilized eggs are transported to hatchery facilities for incubation and rearing. Juveniles are released for supplementation purposes in the Kootenai River, ID


Effective Population Size and Sibship Reconstruction
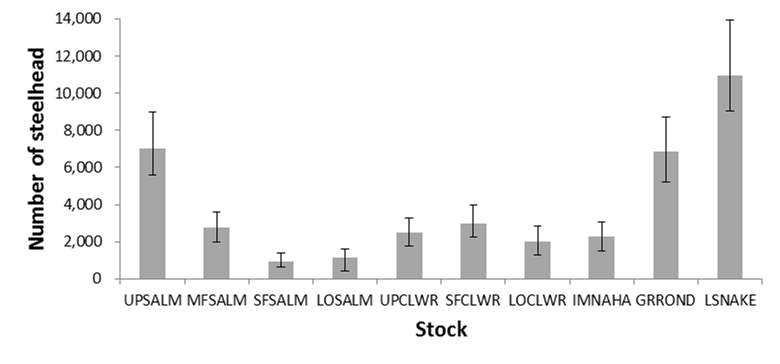
Effective Population Size and Sibship Reconstruction: An evaluation of precision and accuracy using Snake River Hatchery Steelhead
The Knights of NE: Michael W. Ackerman, Jesse D. McCane, Ninh Vu, Craig Steele, Matthew Campbell
INTRODUCTION:
Effective population size (NE) is arguably the most important metric in conservation biology. NE affects population viability, genetic drift, and natural selection and accounts for a population’s abundance and demography. In most natural populations it is difficult to collect adequate data to calculate NE directly. Therefore, genetic methods have been developed to estimate NE. One method is to reconstruct sibling relationships among a sample of offspring using genotype data. Accurate sibship reconstruction allows one to assess variance in reproductive success and facilitates NE estimation. In this study, we evaluated our ability to reconstruct sibling relationships and estimate the effective number of breeders (NB) among Snake River hatchery steelhead stocks with known relationships and known NB.
DATA:
We used genotype data from five hatchery broodstocks genotyped annually for parentage based tagging (PBT) in the Snake River basin (Steele et al. 2013). Specifically, we used data from adults returning to hatcheries in 2012 & 2013 that were offspring of broodstock spawned in 2009 (Table 1). We know the sibling relationships among offspring returning in 2012 & 2013 because they have been assigned parentage to parents spawned in 2009 (many of which are verified by cross records). For this study, we then ignored the parents’ genotype data, because we want to evaluate our ability to perform sibling reconstruction when only offspring data is available. We use 96 O. mykiss SNPs used for PBT in the Columbia River basin. We calculated the true NB of each broodstock using the parentage-without-parents (PWOP) method (Waples & Waples 2011) using genotype data from all returning offspring.
1) What is the accuracy of sibship reconstruction and NB estimation when using all offpsring?
We assessed COLONY’s ability to correctly identify true full siblings, half siblings, and unrelated pairs among all offspring for each hatchery. Analyses were run using both the monogamy (ignores half siblings) and polygamy settings in COLONY. In total, >2,500,000 pairwise sibling relationships were assessed. We further evaluated COLONY’s ability to estimate the true NB. All analyses were repeated using three assumed genotype error rates (0.0001, 0.0005, 0.001).
2) What sample size is necessary to accurately estimate the true NB of each hatchery stock?
We then iteratively sampled the offspring at 10%, 20%,…90% of the total number of offspring for each hatchery. We produced 10 random draws at each sample size resulting in 90 COLONY runs per hatchery (450 total). All runs were performed using the monogamy setting (based on results from #1) and an assumed 0.001 genotype error rate.
RESULTS:
1) What is the accuracy of sibship reconstruction and NB estimation when using all offpsring?
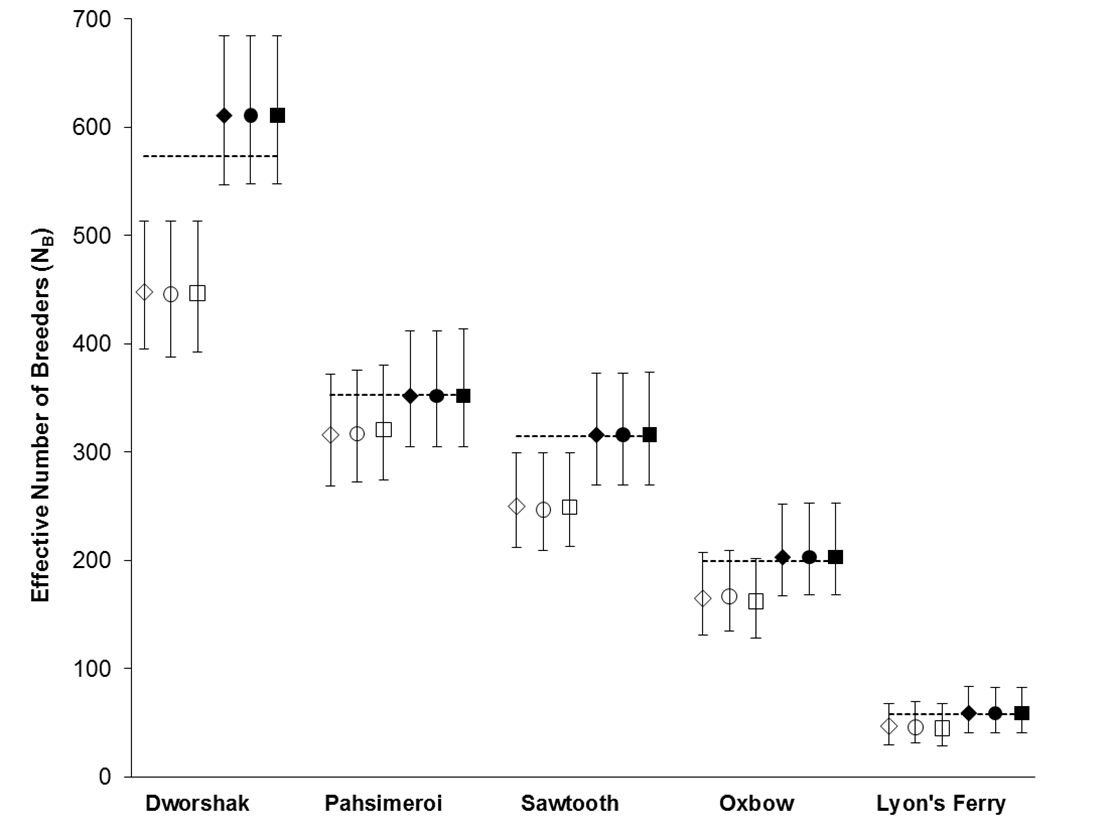
Using the polygamy setting, COLONY correctly identified 95.9% of all true full sibling pairs and 99.8% of the true unrelated pairs. However, only 3.5% of true half sibling pairs were correctly identified. Using the monogamy setting, 99.8% of true full sibling pairs and 100% of unrelated pairs were correctly identified. COLONY accurately estimated the true NB for all 5 hatcheries when assuming a monogamous mating system (Figure 1). Due to poor half sibling reconstruction using 95 SNPs, all analyses for part 2 were performed assuming monogamy.
2) What sample size is necessary to accurately estimate the true NB of each hatchery stock?
For every hatchery, a sample size near or greater than the true NB provided an accurate estimate of NB (Figure 2) and the standard deviation among estimates decreased with increasing sample size.
DISCUSSION:
Our ultimate goal is to accurately estimate the NB and NE of wild populations of Snake River steelhead using a single sample of offspring to assess population viability. Lessons learned in this study will be applied to future research on wild populations: 1) a sample size near or above the true NB is necessary to provide an accurate estimate, and 2) we can iteratively subsample offspring and assess whether we’ve achieved an asymptote in our estimates to know whether we’ve obtained an adequate sample size. Future research is needed to assess the number of SNPs necessary to perform accurate reconstruction of half-sibling pairs.
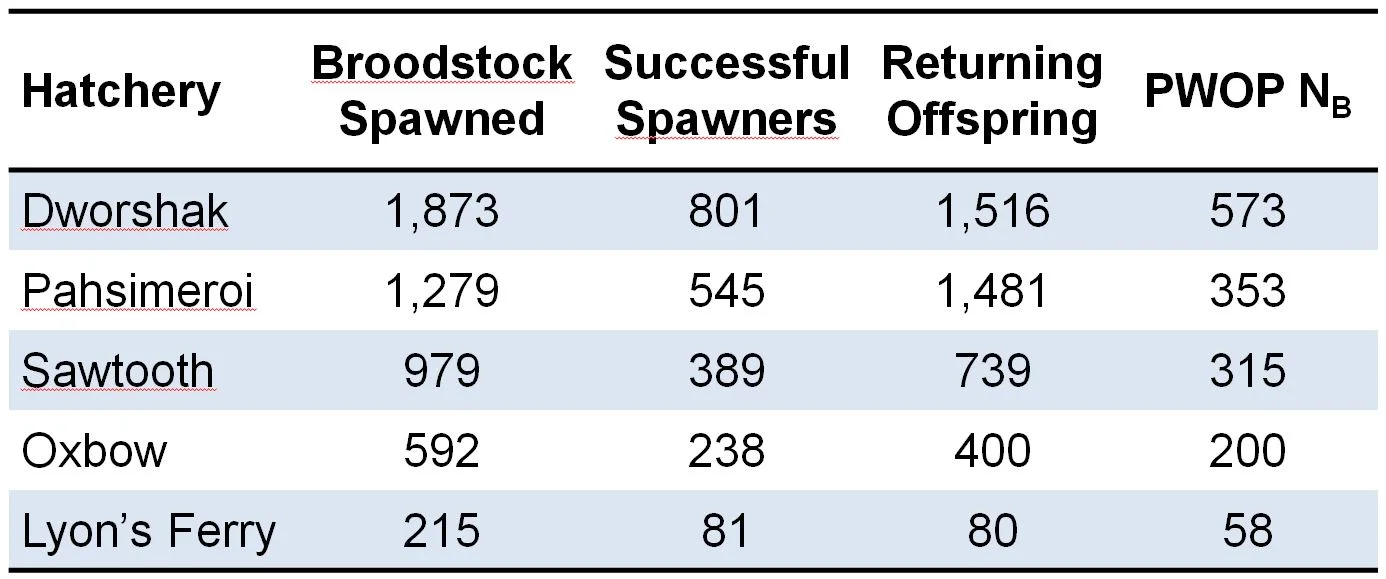
Table 1. Steelhead broodstock spawned at 5 Snake River hatcheries and number of successful spawners and returning offspring used to calculate PWOP NB.
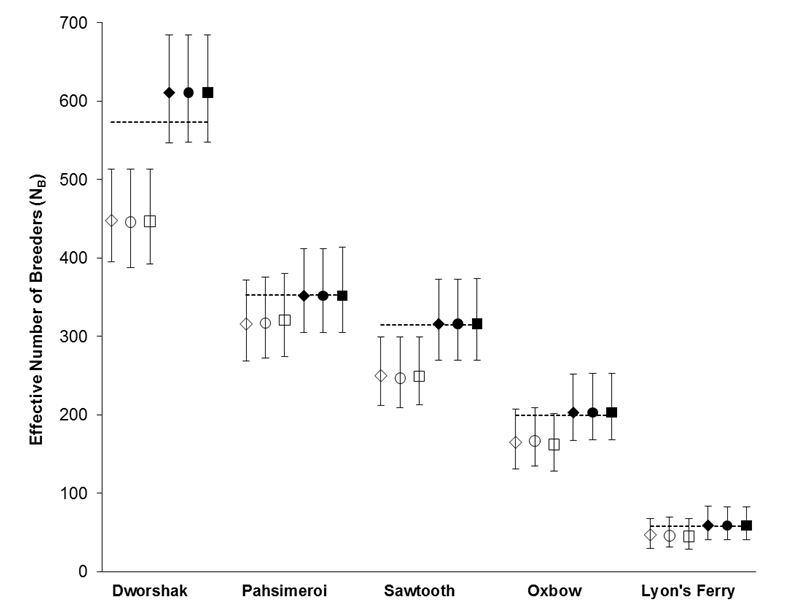
Figure 1. NB estimates for 5 Snake river hatcheries using both the polygamy (open symbols) and monogamy (filled symbols) settings in COLONY. Analyses were repeated using three assumed genotyping error rates: 0.0001 (diamonds), 0.0005 (circles), and 0.001 (squares). Dashed lines represent the true NB estimated using PWOP (Waples and Waples 2011).

Figure 2. Estimates of NB for 4 Snake River hatcheries at intervals of 10%, 20%,…100% of the total number of offspring. Ten random draws of offspring were made at each interval for 10% to 90%. Each dot represents one run of COLONY. Dashed lines represent the true NB. The solid line is the standard deviation among the 10 runs (secondary axis). Lyon’s Ferry is excluded for poster formatting.
Developing SNP markers for Shoshone sculpin
Ninh Vu , Eagle Fish Genetics Lab, Pacific States Marine Fisheries Commission, Boise, ID
Matthew R. Campbell , Eagle Fish Genetics Lab, Idaho Department of Fish and Game, Eagle, ID
Shawn R. Narum , Fish Science, Columbia River Inter-Tribal Fish Commission, Hagerman, ID
Tracy Richter , Idaho Power Company, Boise, ID
John Anderson , Idaho Power Company, Boise, ID
The Shoshone sculpin Cottus greenei is one of eight sculpin species found in Idaho (Simpson and Wallace 1982). Although some of Idaho’s sculpin species are widely distributed throughout the state, Shoshone sculpin are endemic only to springs and spring creeks along a 38–km (24–mile) section of the Snake River in the Hagerman Valley of south–central Idaho (Griffith 1984). Because of its limited distribution and reductions in its spring habitat (Griffith and Kuda 1994), Shoshone sculpin are recognized as a species of special concern by the Idaho Department of Fish and Game (IDFG), and a species of concern by the USFWS (IDCDC 2003).
Recently, Idaho Department of Fish and Game (IDFG) and Idaho Power biologists have collaborated on research projects aimed at better understanding the distribution, life history, and genetic population structure of Shoshone sculpin and to develop genetic monitoring techniques for estimating abundance and long-term viability. A previous survey of these species using 7 microsatellite markers succeeded in describing diversity and structure of the species, but failed to obtain accurate or precise estimates of effective population size (Campbell 2011). The development of SNP genetic markers was recommended:
-To more accurately estimate effective population size
-Provide genetic “tagging” of individual fish for movement and survival studies
-SNP markers can be used for parentage studies if translocation and/or reintroduction efforts are deemed necessary in the future
In this study, we used samples collected from throughout the range of the species (Figure 1) and Restriction site Associated DNA (RAD) sequencing to identify single nucleotide polymorphic (SNP) genetic markers. From this RAD sequencing, we identified 419 SNP markers to describe the initial genetic diversity and differentiation among the 11 sample locations. Results indicated high levels of genetic differentiation among sites (FST = 0.075 - 0.134) and within population genetic diversity (HE) ranging from 25.5 – 32.7%). To display the genetic relationships among populations, a neighbor-joining dendrogram was generated from genetic distances (Cavalli-Sforza and Edwards 1967) between all population (Figure 2).
We developed 96 Taqman SNP assays from the 419 initial markers and screened them on 731 Shoshone sculpin samples collected from Banbury Springs in 2014. Simulations performed on this dataset indicated that this SNP panel has sufficient power for mark-recapture and parentage studies.
We estimated the contemporary effective population size of the Banbury Shoshone sculpin population using the linkage disequilibrium method (Hill 1981) option in NEEstimator v2 (Do 2014). Under a random mating model, the estimated parental generation inbreeding effective size (Neb; effective number of breeding adults) was ~1000 (Figure 3).
In conclusion, RAD sequencing allowed efficient and rapid identification of thousands of variable SNPs in a non-model organism. We developed and optimized a set of Taqman markers that are easily scorable, and highly variable across the species range. Results indicate that this SNP panel will be more powerful for studies involving mark – recapture, parentage, and estimation of effective population size (NE) than microsatellite panel developed previously.
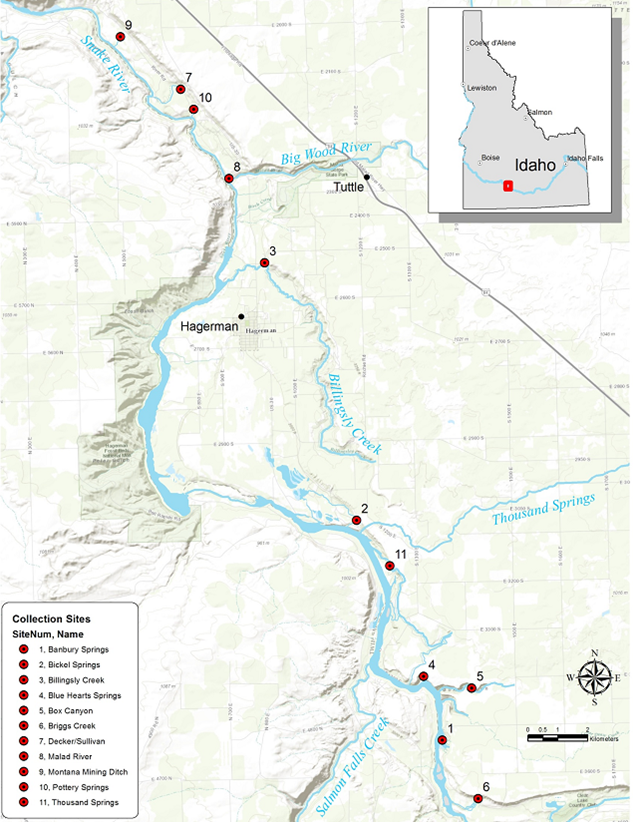
Figure 1. 12 individuals sampled from each of 11 sample locations across the species range in the Hagerman Valley, ID for RAD Sequencing.
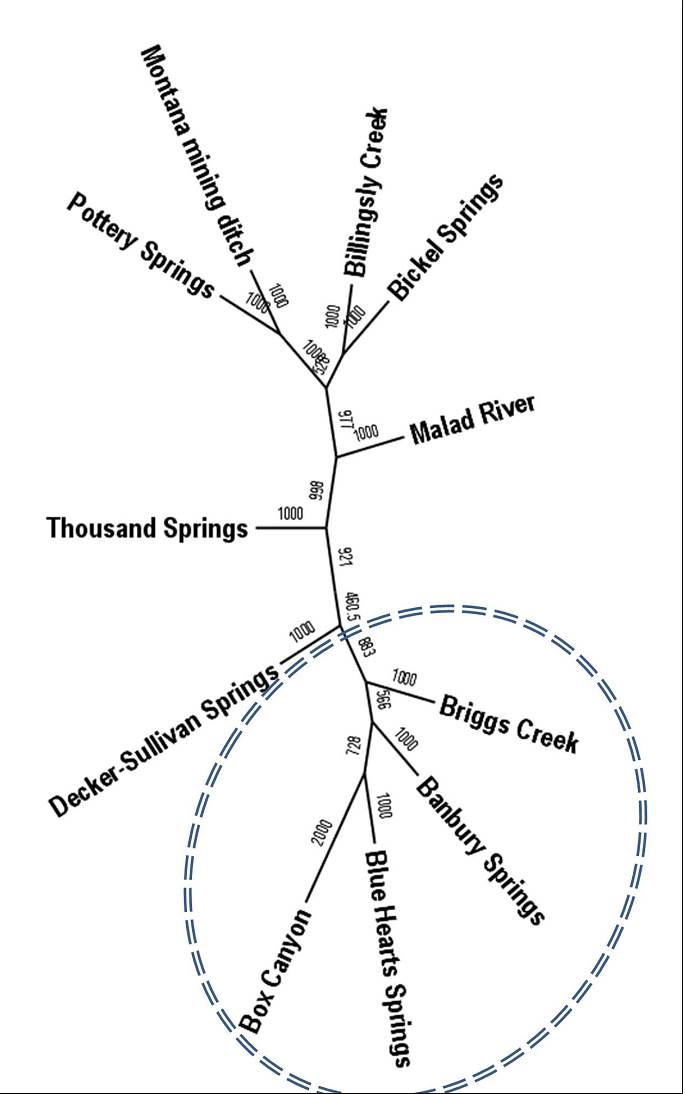
Figure 2. Neighbor-joining dendrogram from genetic distances (Cavalli-Sforza and Edwards 1967) between all populations.
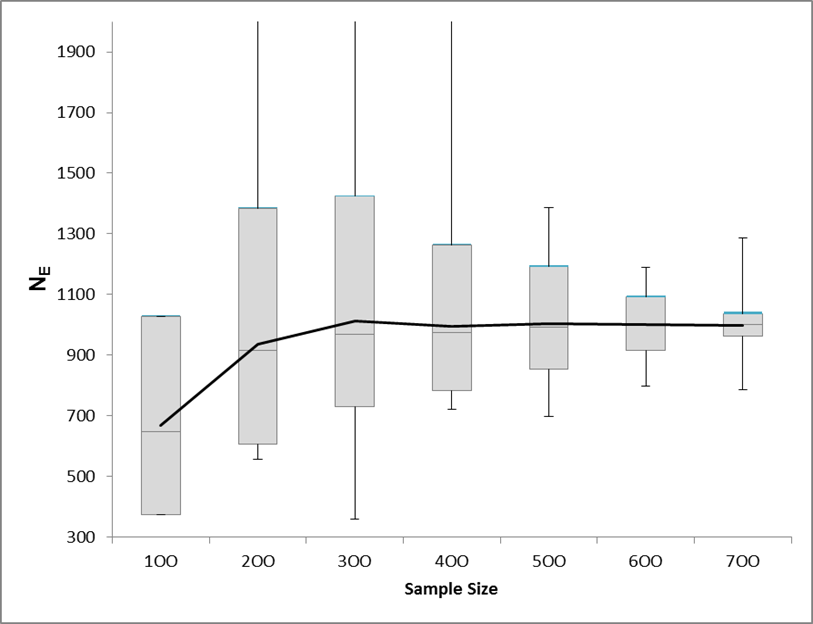
Figure 3. Effective population size estimates from LDNE (with 95% CI) of varying sample sizes (100, 200, 300, 400, 500, 600, and 700) for the Banbury Springs population. NE asymptotes at ~1000.

Funding for this project was provided by the Idaho Power Company.
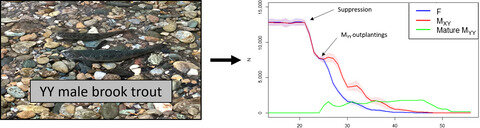
Trojan Y Chromosome Strategy in Brook Trout – Idaho Fish and Game
-Trojan Y Chromosome Strategy in Brook Trout – Idaho Fish and Game
-Super Male brook trout may improve angling
-YY-Male Brook Trout as an Eradication Tool of Wild Brook Trout Populations in New Mexico
-Idaho Continues work on YY Males, an Approach for Eradicating Invasive Fish Populations
By Roger Phillips, Public Information Supervisor
Wednesday, September 14, 2016 - 11:11 AM MDT
Technique won a national award for fisheries restoration
Hatcheries have long been used to replenish and restore fish populations, but can they also be used to reduce or eradicate them? Idaho Fish and Game researchers are studying whether using traditional hatchery technology in a nontraditional way can eliminate unwanted fish populations in the wild.
Fish and Game researchers and hatchery staff are collaborating on a project using 50 year-old technology to develop a monosex fish population whose offspring can only produce males. These males have two YY chromosomes (YY) rather than the usual XY arrangement.
Stocking YY-male hatchery fish into a body of water with an undesired fish population could change the sex ratio to all males within a few generations, and the unwanted fish population would eventually fail to reproduce and therefore die off. Once accomplished, Fish and Game would stop stocking those fish and fisheries managers would then restock that body of water with a more desirable fish species.
Brook trout were selected for the first YY project because they are short lived and quick to sexually mature, which enables researchers to rapidly produce the hatchery broodstock and test the technique in a natural environment. Brook trout are also good candidates because they are nonnative, frequently overpopulate, and stunt in both lakes and streams, which means fish are too small to be of interest for most anglers.
The YY technique begins in a hatchery, where young brook trout are exposed to low-doses of a naturally occurring female hormone, estradiol, which has no effect on female fish, but causes male fish to produce eggs when they mature. The egg-producing males are crossed with standard males, which produce about 25 percent YY-male offspring. Those offspring are used to produce another generation that will theoretically produce exclusively male offspring when bred with any other brook trout.
Brook trout produced in the program for stocking in the wild are not exposed to any hormones and appear like all other brook trout, but they carry two male chromosomes instead of one.
While it sounds complex, it’s a fairly simple method of using hormones to affect gender in a segment of the population, then selectively breeding them to get an entire population to produce one gender. It’s routinely done in commercial aquaculture hatcheries to raise identical-looking food-fish, increase growth rates, and control reproduction.
If the program with brook trout proves successful, the “YY male” method could eradicate or limit other undesirable fish species in select waters, perhaps even large bodies of water with carp infestations, or other unwanted fish that limit game fish populations and harm habitat.
Fish and Game has long used fish toxicants to eradicate unwanted fish from entire bodies of water, but toxicants are typically limited to smaller bodies of water, such as ponds, small lakes and reservoirs or small streams.
Netting, trapping, and other fish removal methods also rid waters of unwanted fish, but those efforts are rarely a long-term solution because a few fish usually escape and spawn successfully. All those methods are time consuming, labor intensive, and often have to be repeated years later when unwanted fish populations rebuild.
Fish and Game officials hope the YY-male approach will be a cost-effective technique to control undesirable fish populations. Gary Byrne, the Fish Production Manger overseeing the hatchery portion of Idaho’s YY brook trout program, said it only took four years to develop the YY brook trout broodstock.
Head Fisheries Researcher Dan Schill, who led the team conducting the research project, said they are encouraged by the low cost of broodstock development, and they hope the technique will curb brook trout populations in waters where it’s being tested.
“The proof will be in the pudding over the next few years when our research staff obtain results confirming whether stocked YY fish successfully spawn in the wild and are ultimately effective in reducing the percentage of wild female brook trout in test waters,” Schill said.
Stocking trials of YY Brook Trout in four Idaho streams began in 2014, and the first results are encouraging. A marked YY Male was observed actively spawning in October with a wild female, and testing done on wild fry in study streams in 2015 conclusively showed that some YY males successfully spawned. Of equal note, all progeny of stocked YY fish found were XY males, exactly as predicted and as investigators hoped.
Fish and Game officials presented their findings at the August 2016 American Fisheries Society (AFS) national meeting in Kansas City, which has generated excitement in the fisheries science community. There, the AFS announced Fish and Game’s YY Male Brook Trout Research Program won the 2016 Sport Fish Restoration Outstanding Project award in the category of Research and Surveys.
The awards highlight the importance and effectiveness of the sport fish restoration program and recognize excellence in fisheries management, research and education.
Questions and answers about the YY-male fish program
Q: I understand the basic method of producing YY males in the hatchery, but how does the process of eliminating an undesirable fish population work in an actual stream or lake?
A: In natural fish populations, females have two X sex chromosomes (XX) and males have an X and Y chromosome (XY). When two wild fish spawn, offspring can only inherit one sex chromosome from each parent. Offspring receive an X from the female because that’s all she produces. Half the progeny receive an X from the male side and half receive a Y, which produces a 50:50 sex ratio.
When a hatchery-produced YY male spawns with a wild, XX female, all the progeny inherit one Y from the male and one X from the female and therefore would all be XY males. The basic idea is to continue stocking YY Males into the wild population until all the fish in the water are male, then stocking would end and the population would consequently die off.
Q: Is Fish and Game trying to eradicate all brook trout?
A: No, that’s not the intent of this program. While there is interest in eliminating brook trout in specific waters where they severely impact native species or stunt and become undesirable to anglers, this is primarily an effort to test the basic YY male concept in small, isolated waters. Anglers will continue to find hundreds of streams and mountain lakes in Idaho with wild Brook Trout populations.
Q: Are these brook trout being stocked considered a GMO?
A: No. Because no genes are “spliced” into the target fish genome from another fish species, the YY male fish produced are not genetically modified organisms (or GMO’s), a plus that has been noted by the authors of several recent scientific papers reviewing the YY male approach.
Q: What happens if someone eats a fish that’s been exposed to estradiol.
A: Fish exposed to the hormone in the program remain in enclosed hatchery production silos and are never stocked so it’s virtually impossible for that to occur. However, the doses given to tiny fry are very low and the 100 percent clearance rate from tissues is a matter of days, well over a year before fish become a size of interest to anglers. For these reasons, it is inconceivable that an angler or other animals would be exposed to estradiol, which is a common and frequently used human prescription drug that’s also used in aquaculture.
Q: Could the YY approach lead to eliminating other types of unwanted, non-native, or nuisance fish?
A: Although not the goal of the current program, Fish and Game is always trying to find ways to improve fishing and water quality. Where an undesirable species is limiting fishing opportunity, this is one method that could be attempted if the present experiments on Brook Trout prove successful.
Q: Is it reversible if for some reason you wanted that fish back?
A: Yes. All Fish and Game would have to do is stop stocking YY fish and the population would return to a normal 50:50 sex ratio.
Q: How long would it take for the population to go all male?
Preliminarily, it looks like in the case of brook trout in streams, somewhere between four and 10 years will be the answer depending on number of fish stocked, etc. There are scenarios where it might take longer, but in those instances, it is unlikely that such an approach would be deployed. F&G biologists are in the process of submitting work on this exact question for publication in a fisheries journal where it will receive careful peer review.
Q: Why wouldn't you allow anglers to reduce unwanted fish populations?
A: We generally employ high bag limits (or remove bag limits all together) to encourage angler harvest on undesirable fish species. These new techniques won't replace that. Netting, trapping, and other fish removal methods can also be used to rid waters of unwanted fish, but those efforts are aren't silver bullets because a few fish can escape and spawn successfully. All those methods are time consuming, labor intensive, and may need to be repeated years later when unwanted fish populations rebuild. This new approach may make it impossible for populations to rebuild after they are suppressed.



Interspecific hybridization in a large‐river population of YCT: A 20‐year evaluation
Hybridization between native and nonnative fishes represents a global threat to biodiversity. Understanding how hybridization changes in response to management actions is critical to evaluating the efficacy of conservation efforts.
Methods
We quantified changes in levels of hybridization between Yellowstone Cutthroat Trout Oncorhynchus virginalis bouvieri and Rainbow Trout Oncorhynchus mykiss in the South Fork Snake River watershed, where a multipronged approach has been implemented to protect the evolutionary distinctiveness of one of the last remaining large‐river populations of Yellowstone Cutthroat Trout.
Result
Over a 20‐year period, we observed an increase in the number of sample reaches without hybrids in the South Fork Snake River watershed; however, contrasting patterns were noted in main‐stem and tributary reaches. Through time, hybrid abundance increased at main‐stem reaches of the South Fork Snake River below Palisades Dam but decreased in tributaries. Efforts to reduce hybridization in spawning tributaries, including both suppression and selective passage weirs, were effective at preventing the expansion of hybridization in resident and migratory populations. Multimodel inference was used to understand factors affecting levels of hybridization, and year, sampling reach, and the interaction thereof was identified as the best‐fit model but explained only a small percentage of the overall variation, suggesting that other factors not captured in our model were driving patterns in hybridization.
Conclusion
Changes in hybridization in the South Fork Snake River watershed are likely the result of multiple processes, namely management actions to reduce Rainbow Trout and hybrids in tributaries, as well as demographic changes in Rainbow Trout in the main‐stem river below Palisades Dam. Our results suggest that Yellowstone Cutthroat Trout populations in the South Fork Snake River watershed have not experienced widespread interspecific hybridization with Rainbow Trout but that proactive management will be necessary to ensure long‐term conservation.


Figure 1.
A map of the South Fork Snake River watershed and associated tributaries in portions of Idaho and Wyoming where Yellowstone Cutthroat Trout were sampled for genetic analysis. Within each creek (except Fall Creek), three sections were sampled corresponding to lower, middle, and upper reaches of the tributary. Sampling reaches on the main stem of rivers include Lorenzo and Conant on the South Fork Snake River and Narrows Pass on the Salt River, a tributary to the South Fork Snake River. Selective passage weirs operated on spawning tributaries are shown as is the placement of Palisades Dam. The arrow denotes the direction of stream flow.

Figure 2.
The percentage of fish identified as hybrid (i.e., contained at least one species‐diagnostic SNP for Rainbow Trout) from tributaries to the South Fork Snake River, Idaho. Estimates were generated by pooling fish across tributaries.