Developing SNP markers for Shoshone sculpin
Ninh Vu , Eagle Fish Genetics Lab, Pacific States Marine Fisheries Commission, Boise, ID
Matthew R. Campbell , Eagle Fish Genetics Lab, Idaho Department of Fish and Game, Eagle, ID
Shawn R. Narum , Fish Science, Columbia River Inter-Tribal Fish Commission, Hagerman, ID
Tracy Richter , Idaho Power Company, Boise, ID
John Anderson , Idaho Power Company, Boise, ID
The Shoshone sculpin Cottus greenei is one of eight sculpin species found in Idaho (Simpson and Wallace 1982). Although some of Idaho’s sculpin species are widely distributed throughout the state, Shoshone sculpin are endemic only to springs and spring creeks along a 38–km (24–mile) section of the Snake River in the Hagerman Valley of south–central Idaho (Griffith 1984). Because of its limited distribution and reductions in its spring habitat (Griffith and Kuda 1994), Shoshone sculpin are recognized as a species of special concern by the Idaho Department of Fish and Game (IDFG), and a species of concern by the USFWS (IDCDC 2003).
Recently, Idaho Department of Fish and Game (IDFG) and Idaho Power biologists have collaborated on research projects aimed at better understanding the distribution, life history, and genetic population structure of Shoshone sculpin and to develop genetic monitoring techniques for estimating abundance and long-term viability. A previous survey of these species using 7 microsatellite markers succeeded in describing diversity and structure of the species, but failed to obtain accurate or precise estimates of effective population size (Campbell 2011). The development of SNP genetic markers was recommended:
-To more accurately estimate effective population size
-Provide genetic “tagging” of individual fish for movement and survival studies
-SNP markers can be used for parentage studies if translocation and/or reintroduction efforts are deemed necessary in the future
In this study, we used samples collected from throughout the range of the species (Figure 1) and Restriction site Associated DNA (RAD) sequencing to identify single nucleotide polymorphic (SNP) genetic markers. From this RAD sequencing, we identified 419 SNP markers to describe the initial genetic diversity and differentiation among the 11 sample locations. Results indicated high levels of genetic differentiation among sites (FST = 0.075 - 0.134) and within population genetic diversity (HE) ranging from 25.5 – 32.7%). To display the genetic relationships among populations, a neighbor-joining dendrogram was generated from genetic distances (Cavalli-Sforza and Edwards 1967) between all population (Figure 2).
We developed 96 Taqman SNP assays from the 419 initial markers and screened them on 731 Shoshone sculpin samples collected from Banbury Springs in 2014. Simulations performed on this dataset indicated that this SNP panel has sufficient power for mark-recapture and parentage studies.
We estimated the contemporary effective population size of the Banbury Shoshone sculpin population using the linkage disequilibrium method (Hill 1981) option in NEEstimator v2 (Do 2014). Under a random mating model, the estimated parental generation inbreeding effective size (Neb; effective number of breeding adults) was ~1000 (Figure 3).
In conclusion, RAD sequencing allowed efficient and rapid identification of thousands of variable SNPs in a non-model organism. We developed and optimized a set of Taqman markers that are easily scorable, and highly variable across the species range. Results indicate that this SNP panel will be more powerful for studies involving mark – recapture, parentage, and estimation of effective population size (NE) than microsatellite panel developed previously.
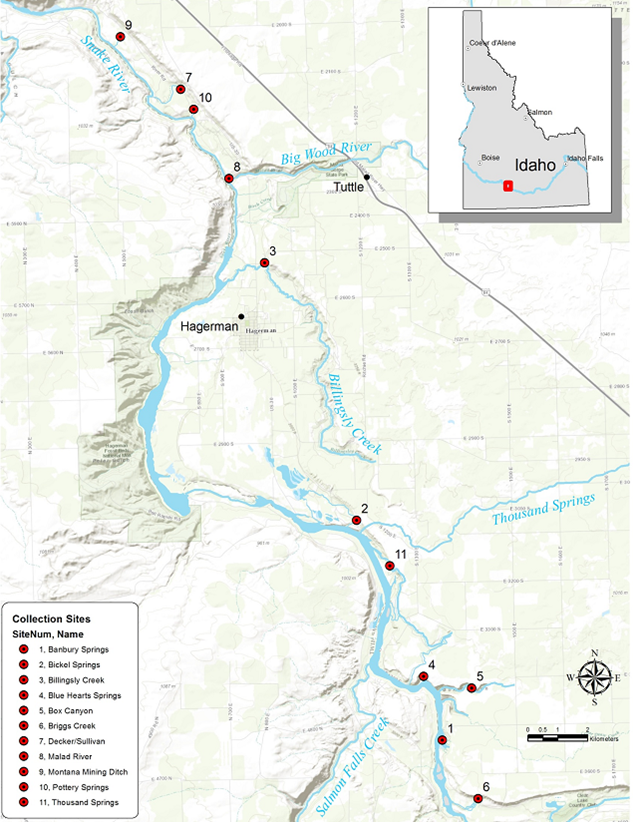
Figure 1. 12 individuals sampled from each of 11 sample locations across the species range in the Hagerman Valley, ID for RAD Sequencing.
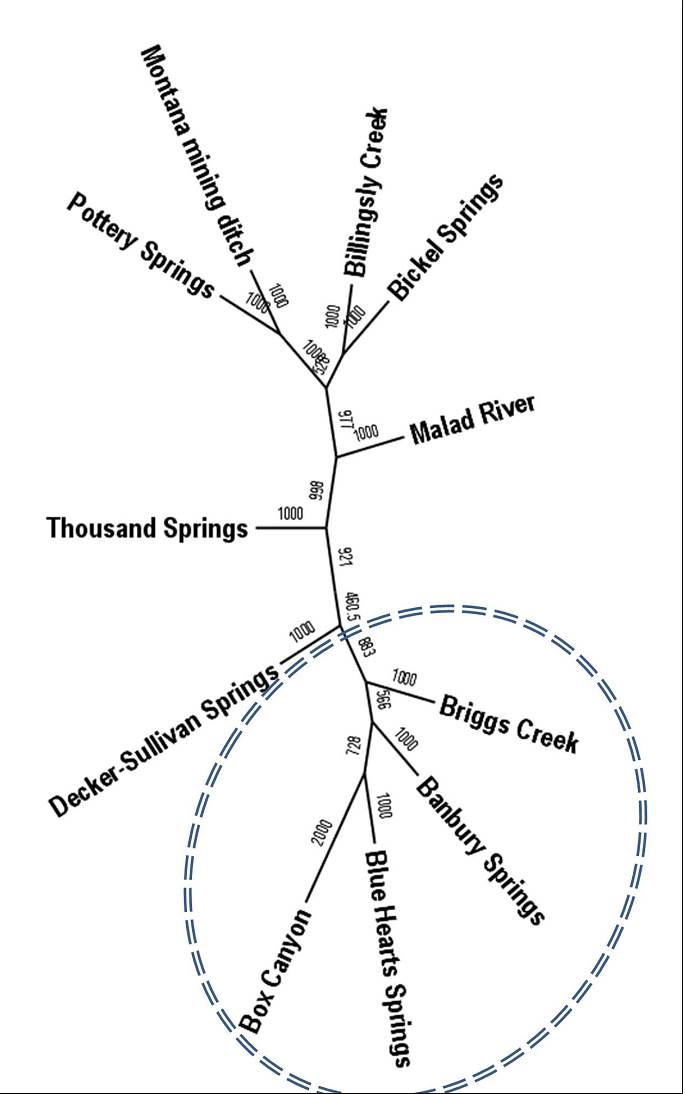
Figure 2. Neighbor-joining dendrogram from genetic distances (Cavalli-Sforza and Edwards 1967) between all populations.
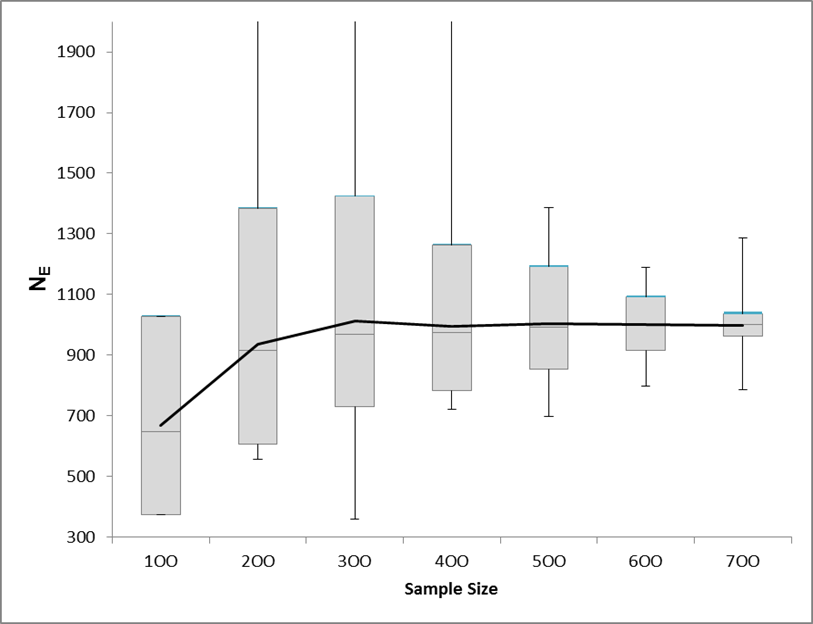
Figure 3. Effective population size estimates from LDNE (with 95% CI) of varying sample sizes (100, 200, 300, 400, 500, 600, and 700) for the Banbury Springs population. NE asymptotes at ~1000.

Funding for this project was provided by the Idaho Power Company.
