Effective Population Size and Sibship Reconstruction
Effective Population Size and Sibship Reconstruction: An evaluation of precision and accuracy using Snake River Hatchery Steelhead
The Knights of NE: Michael W. Ackerman, Jesse D. McCane, Ninh Vu, Craig Steele, Matthew Campbell
INTRODUCTION:
Effective population size (NE) is arguably the most important metric in conservation biology. NE affects population viability, genetic drift, and natural selection and accounts for a population’s abundance and demography. In most natural populations it is difficult to collect adequate data to calculate NE directly. Therefore, genetic methods have been developed to estimate NE. One method is to reconstruct sibling relationships among a sample of offspring using genotype data. Accurate sibship reconstruction allows one to assess variance in reproductive success and facilitates NE estimation. In this study, we evaluated our ability to reconstruct sibling relationships and estimate the effective number of breeders (NB) among Snake River hatchery steelhead stocks with known relationships and known NB.
DATA:
We used genotype data from five hatchery broodstocks genotyped annually for parentage based tagging (PBT) in the Snake River basin (Steele et al. 2013). Specifically, we used data from adults returning to hatcheries in 2012 & 2013 that were offspring of broodstock spawned in 2009 (Table 1). We know the sibling relationships among offspring returning in 2012 & 2013 because they have been assigned parentage to parents spawned in 2009 (many of which are verified by cross records). For this study, we then ignored the parents’ genotype data, because we want to evaluate our ability to perform sibling reconstruction when only offspring data is available. We use 96 O. mykiss SNPs used for PBT in the Columbia River basin. We calculated the true NB of each broodstock using the parentage-without-parents (PWOP) method (Waples & Waples 2011) using genotype data from all returning offspring.
1) What is the accuracy of sibship reconstruction and NB estimation when using all offpsring?
We assessed COLONY’s ability to correctly identify true full siblings, half siblings, and unrelated pairs among all offspring for each hatchery. Analyses were run using both the monogamy (ignores half siblings) and polygamy settings in COLONY. In total, >2,500,000 pairwise sibling relationships were assessed. We further evaluated COLONY’s ability to estimate the true NB. All analyses were repeated using three assumed genotype error rates (0.0001, 0.0005, 0.001).
2) What sample size is necessary to accurately estimate the true NB of each hatchery stock?
We then iteratively sampled the offspring at 10%, 20%,…90% of the total number of offspring for each hatchery. We produced 10 random draws at each sample size resulting in 90 COLONY runs per hatchery (450 total). All runs were performed using the monogamy setting (based on results from #1) and an assumed 0.001 genotype error rate.
RESULTS:
1) What is the accuracy of sibship reconstruction and NB estimation when using all offpsring?
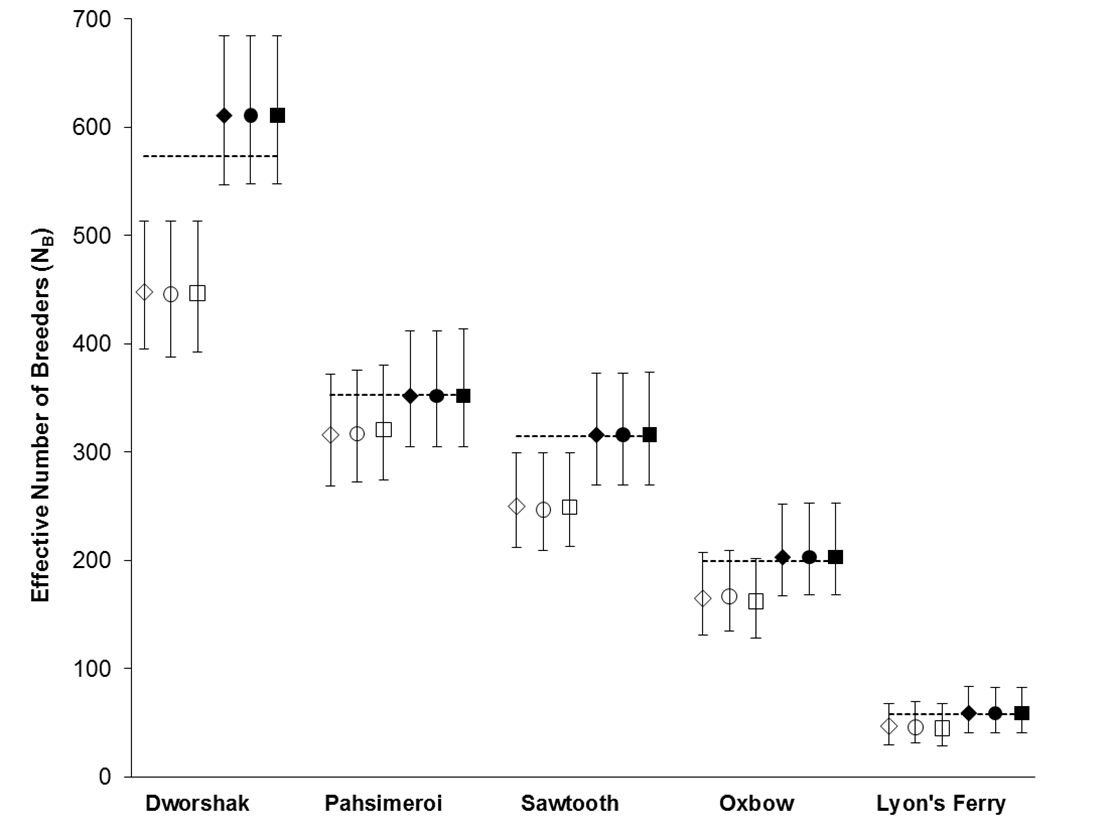
Using the polygamy setting, COLONY correctly identified 95.9% of all true full sibling pairs and 99.8% of the true unrelated pairs. However, only 3.5% of true half sibling pairs were correctly identified. Using the monogamy setting, 99.8% of true full sibling pairs and 100% of unrelated pairs were correctly identified. COLONY accurately estimated the true NB for all 5 hatcheries when assuming a monogamous mating system (Figure 1). Due to poor half sibling reconstruction using 95 SNPs, all analyses for part 2 were performed assuming monogamy.
2) What sample size is necessary to accurately estimate the true NB of each hatchery stock?
For every hatchery, a sample size near or greater than the true NB provided an accurate estimate of NB (Figure 2) and the standard deviation among estimates decreased with increasing sample size.
DISCUSSION:
Our ultimate goal is to accurately estimate the NB and NE of wild populations of Snake River steelhead using a single sample of offspring to assess population viability. Lessons learned in this study will be applied to future research on wild populations: 1) a sample size near or above the true NB is necessary to provide an accurate estimate, and 2) we can iteratively subsample offspring and assess whether we’ve achieved an asymptote in our estimates to know whether we’ve obtained an adequate sample size. Future research is needed to assess the number of SNPs necessary to perform accurate reconstruction of half-sibling pairs.
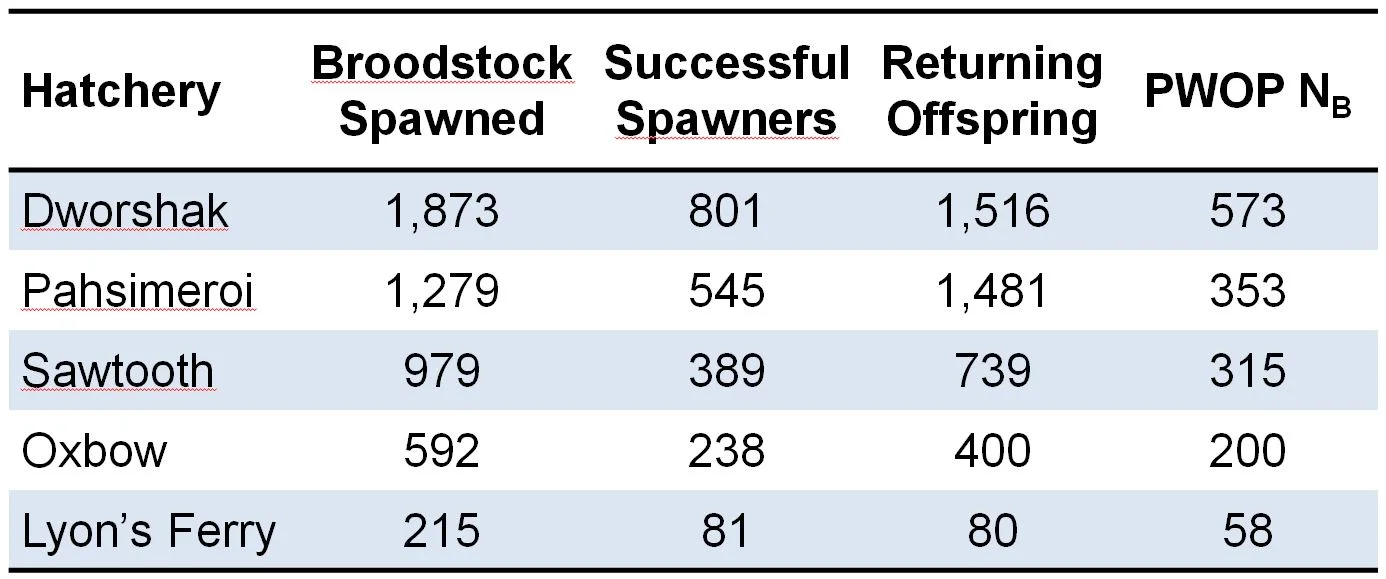
Table 1. Steelhead broodstock spawned at 5 Snake River hatcheries and number of successful spawners and returning offspring used to calculate PWOP NB.
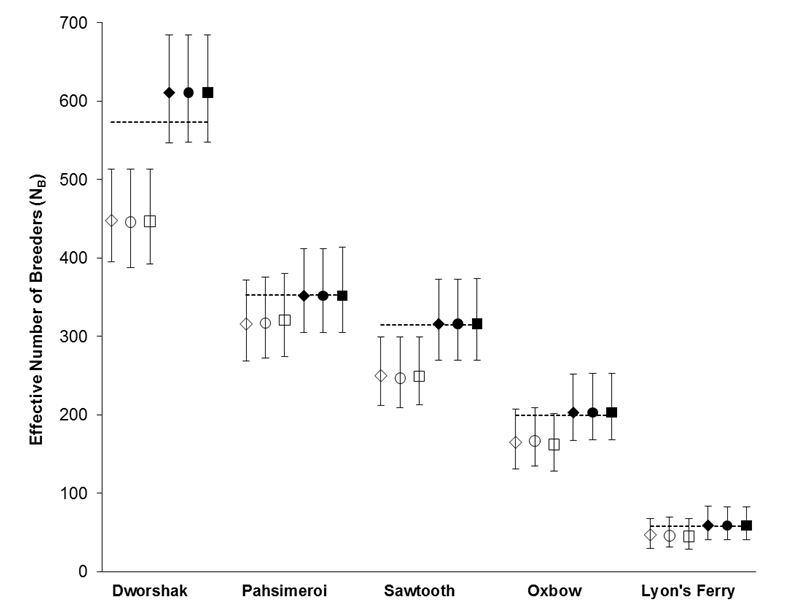
Figure 1. NB estimates for 5 Snake river hatcheries using both the polygamy (open symbols) and monogamy (filled symbols) settings in COLONY. Analyses were repeated using three assumed genotyping error rates: 0.0001 (diamonds), 0.0005 (circles), and 0.001 (squares). Dashed lines represent the true NB estimated using PWOP (Waples and Waples 2011).

Figure 2. Estimates of NB for 4 Snake River hatcheries at intervals of 10%, 20%,…100% of the total number of offspring. Ten random draws of offspring were made at each interval for 10% to 90%. Each dot represents one run of COLONY. Dashed lines represent the true NB. The solid line is the standard deviation among the 10 runs (secondary axis). Lyon’s Ferry is excluded for poster formatting.