Identification of SNPs for PBT of Burbot Lota Lota Using RAD Sequencing
Identification of SNP Markers for Parentage Based Tagging of Burbot Lota Lota Using Restriction-Site-Associated DNA Sequencing
Matthew R. Campbell1, Ryan Hardy1, Amanda Matala2 and Shawn R. Narum2, (1) Idaho Department of Fish and Game, (2) Columbia River Inter-Tribal Fish Commission
Project Summary:
The trans-boundary (Idaho, USA and British Columbia, Canada) population of burbot lota lota native to the Kootenai River once provided a popular sport and commercial fishery, and is culturally significant to the Kootenai Tribe of Idaho. However, the population has experienced significant declines over the last 30 years, due primarily to habitat loss and alteration caused by water storage and diversion. By the late 1990’s, the population was considered functionally extinct, with estimates of fewer than 50 burbot in the wild and little to no recruitment, prompting an on-going international recovery effort. As part of these recovery efforts, managers have been actively developing a hatchery supplementation program to re-build the population and support future tribal sustenance harvest and recreational fisheries.
Each February, spawning adult burbot from Moyie Lake, B.C. are captured via angling, genetically sampled, and spawned on site (Figure 1). Fertilized eggs are transported to hatchery facilities for incubation and rearing. Juveniles are released for supplementation purposes in the Kootenai River, ID (Figure 2). While supplementation breeding programs have the potential to rapidly rebuild depleted natural populations, careful genetic management is critical. In order to monitor genetic diversity and potential inbreeding in the broodstock and to provide Parentage Based Tagging (PBT) of supplementation offspring, a powerful set of new genetic markers were needed. For this project we chose to utilize Restriction site associated DNA sequencing (RADSeq). This is a relatively new and cost-efficient technology allows the rapid discovery of thousands of SNP markers in species that have not been extensively studied previously or for which little existing DNA sequence data exists.
Using RADSeq technology, we discovered 6,517 SNPs and successfully genotyped 1,690 SNPs variable in Moyie Lake samples (Figure 3). The 1,690 SNPs were ranked by MAF (diversity) and HWE and 100 SNPs were submitted for assay-by-design (taqman genotyping assays; Figure 4). A total of 93 SNP assays identified for further genotyping. These exhibited ~44% HE in samples from Moyie Lake. We demonstrated the accuracy of our SNP set for parentage by running a known parent/offspring dataset (N = 344/313, respectively). All 313 offspring accurately assigned to the correct parent pair.
The SNP marker set provides a powerful new tool for managing broodstock and for monitoring and genetically tagging Burbot to track growth, post-release survival and movement of released individuals. In spawn year 2015, no crosses among full-siblings were detected and the effective population size of the spawning population was estimated as 397 (95% C.I. = 260 – 729). A total of 593 burbot adults were sampled from the Kootenai River in 2015. These samples were assigned back to a parental baseline consisting of all Moyie Lake parental broodstock spawned in 2011-2014 (N = 344). Of the 593 adults, we successfully assigned 497 (83.8%) back to the supplementation program (BY2011 = 377, BY2012 = 106, BY2013 = 14).

Figure 1. Spawning adult burbot lota lota from Moyie Lake, B.C.
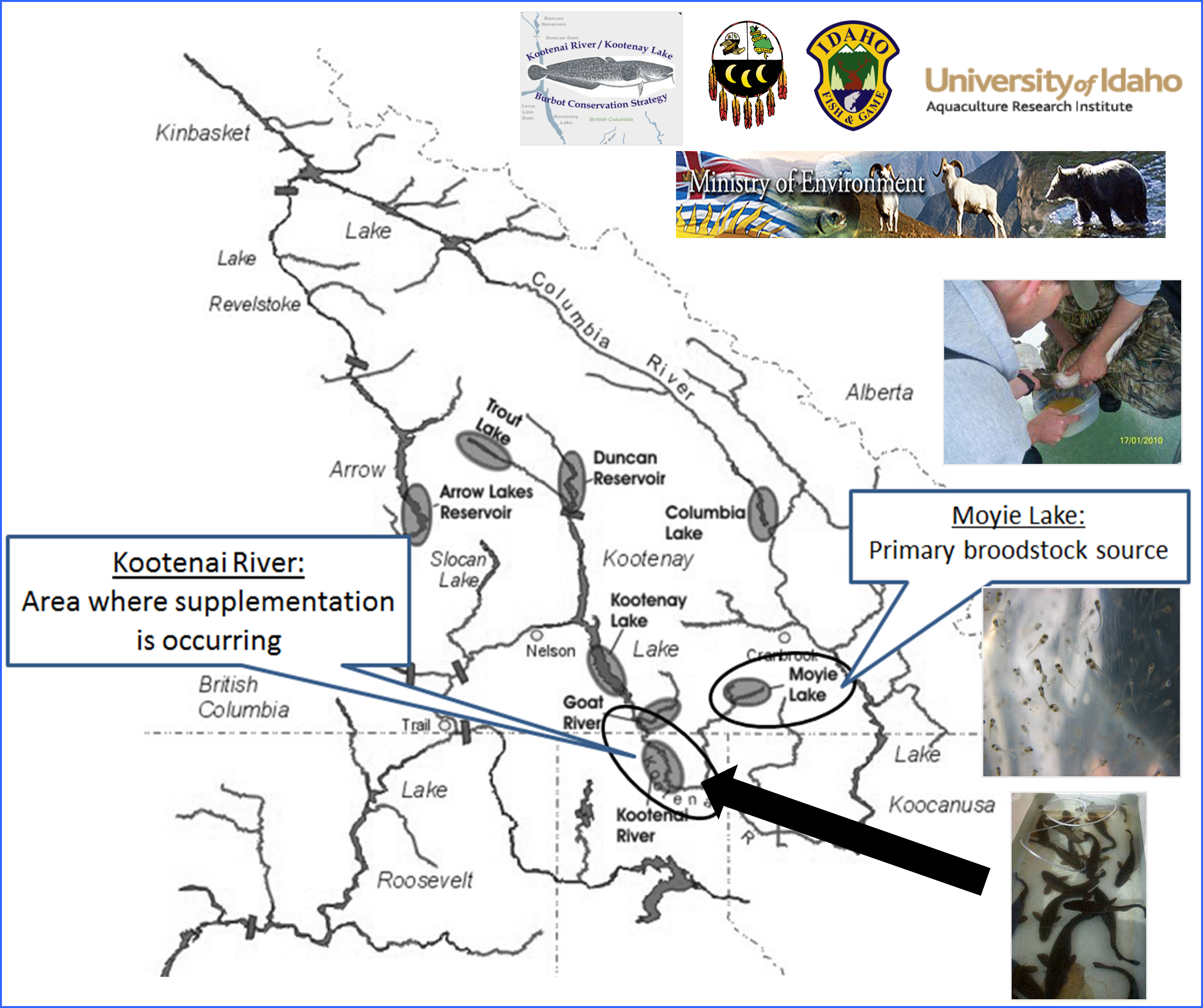
Figure 2.
Each February, spawning adult burbot from Moyie Lake, B.C. are captured via angling, genetically sampled, and spawned on site. Fertilized eggs are transported to hatchery facilities for incubation and rearing. Juveniles are released for supplementation purposes in the Kootenai River, ID
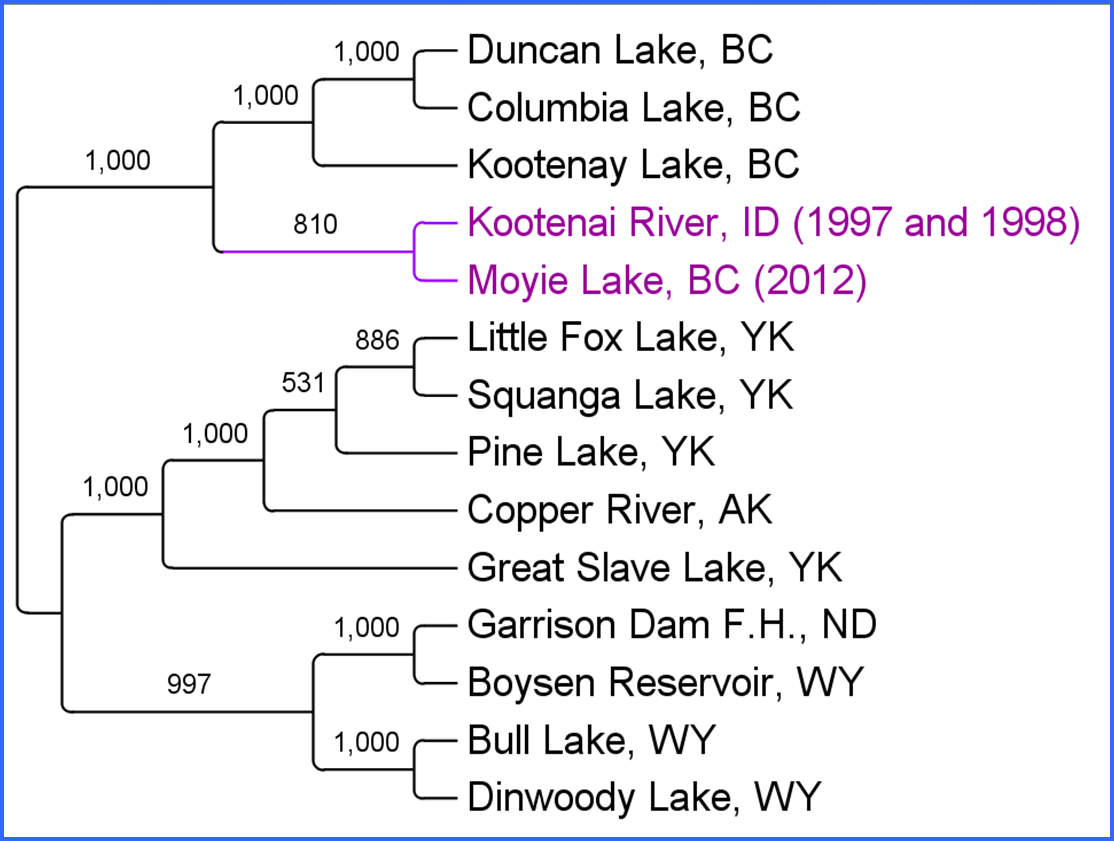
Figure 3. Dendrogram based on 1,690 SNPs showing population relationships among sampled burbot populations. Bootstrap values are reported as the number of the total.
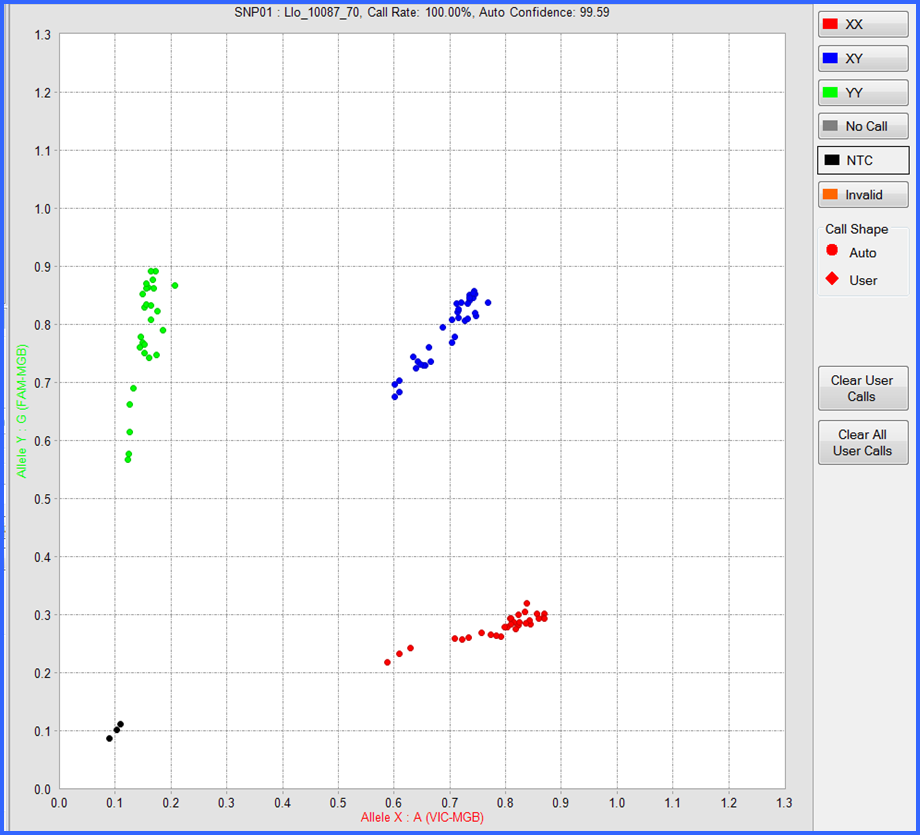
Figure 4. Example cluster plot of 93 Shoshone sculpin samples for SNP36; Cgr_61009 by Taqman assay using the Fludigim EP1 genotyping platform.

Funding for this project was provided by the Bonneville Power Administration.
